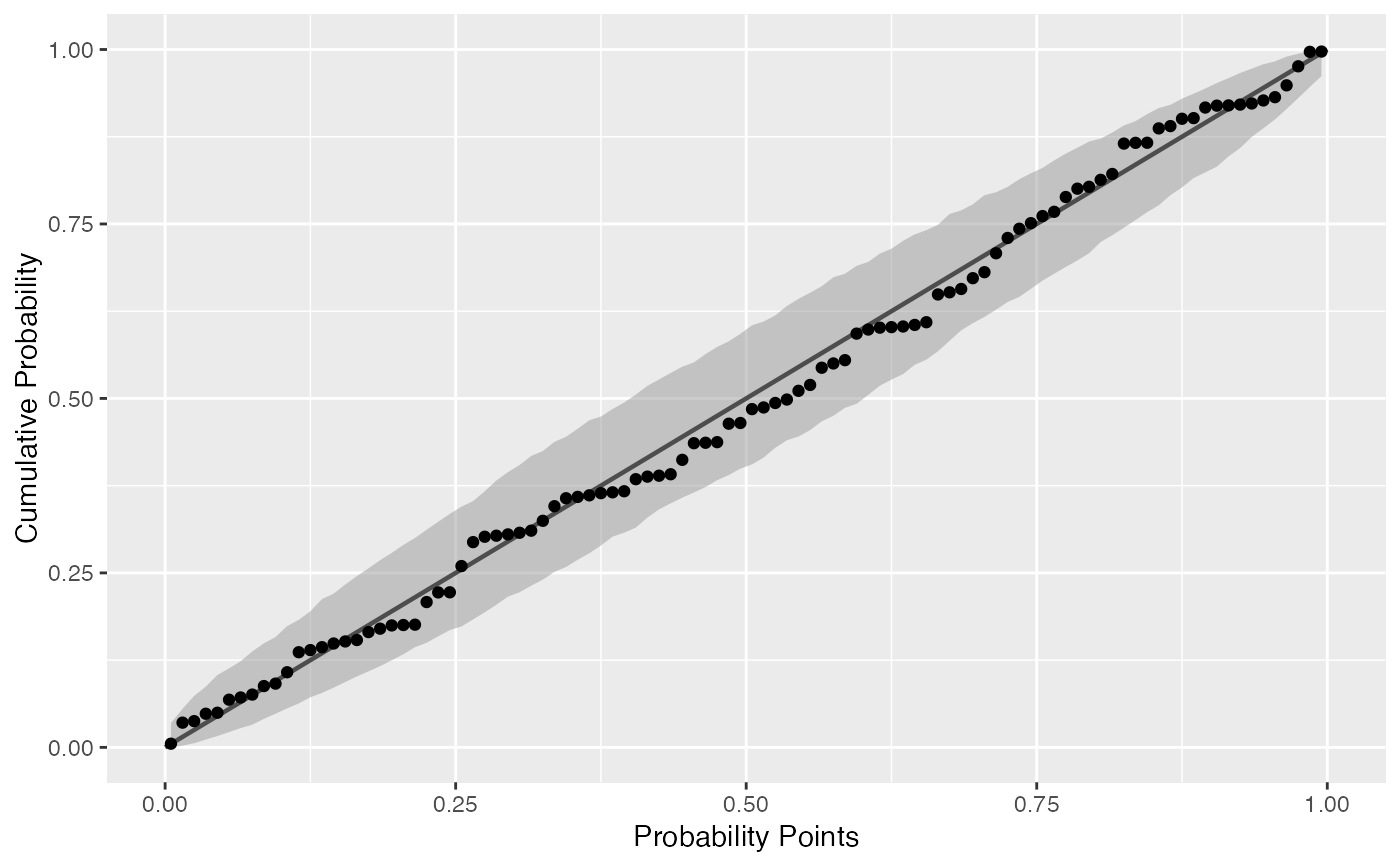
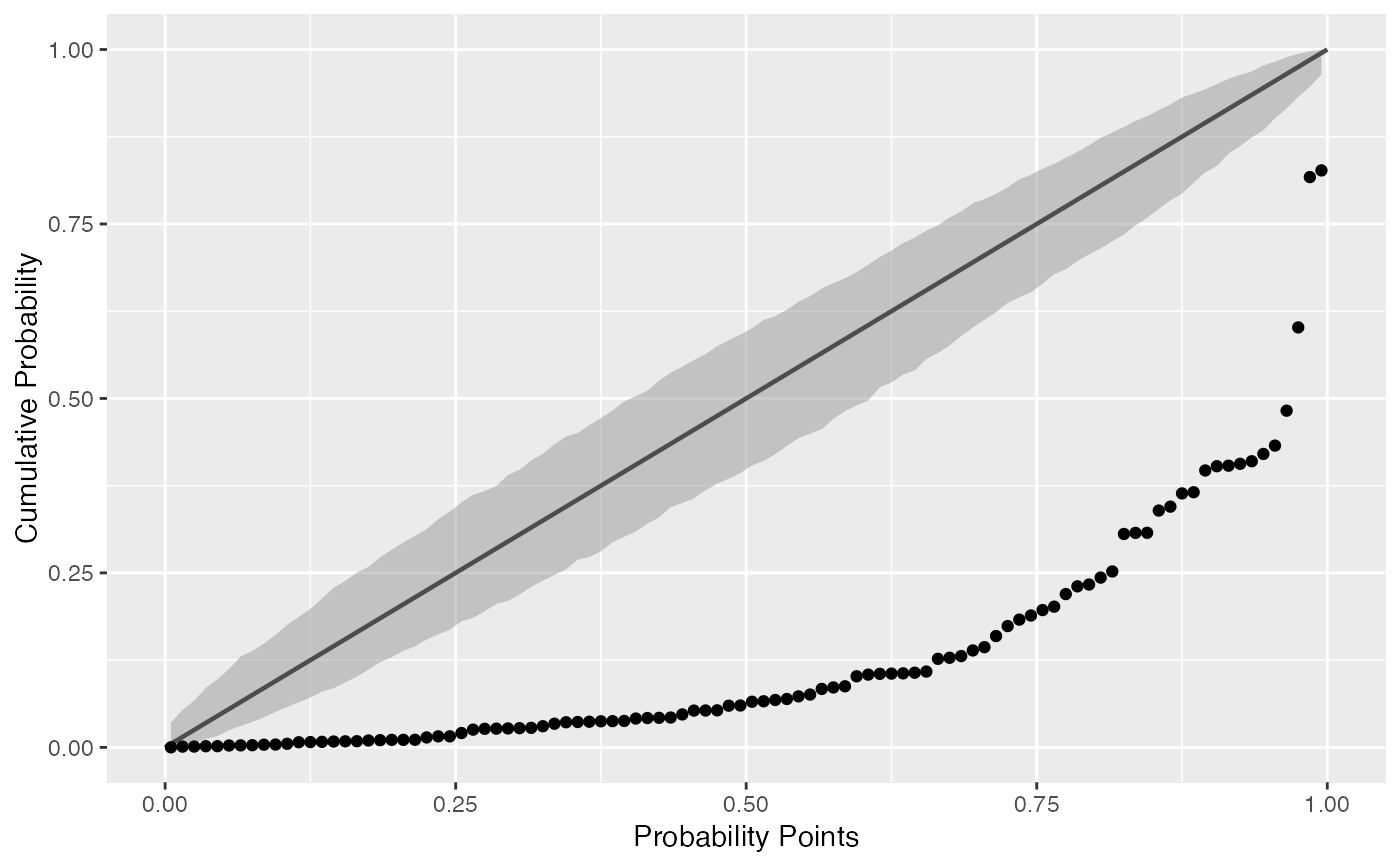
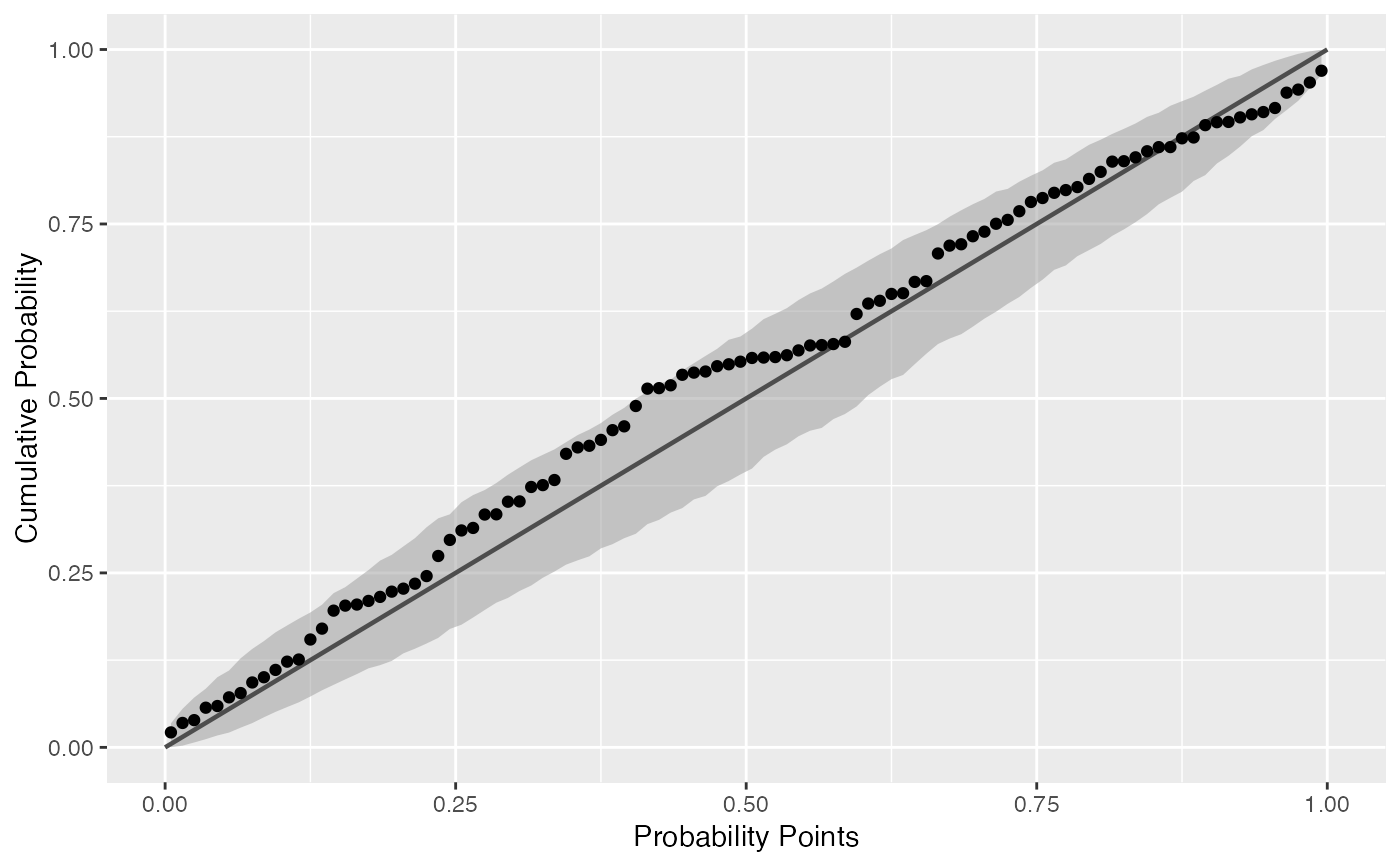
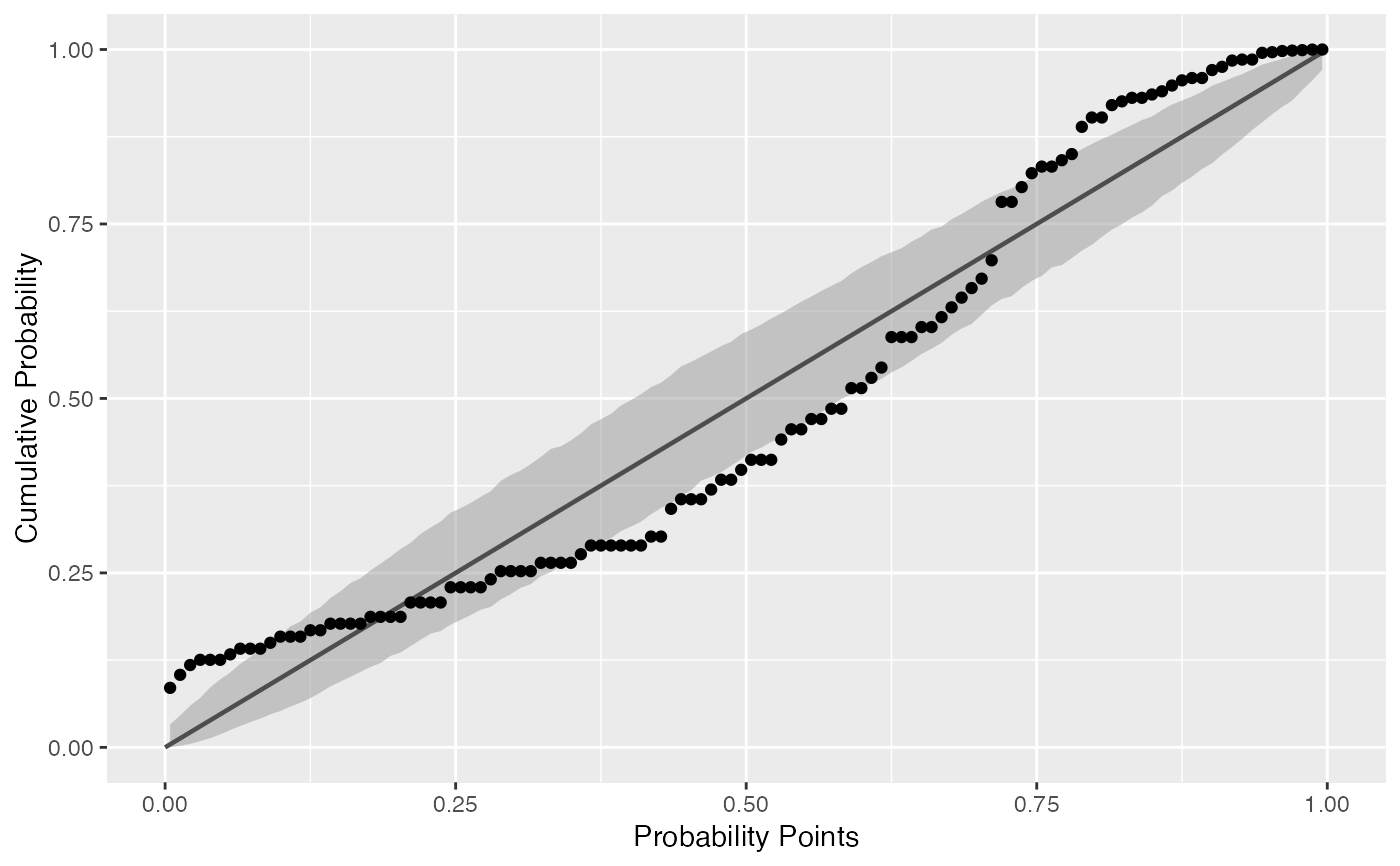
Draws probability-probability confidence bands.
stat_pp_band( mapping = NULL, data = NULL, geom = "ribbon", position = "identity", na.rm = TRUE, show.legend = NA, inherit.aes = TRUE, distribution = "norm", dparams = list(), bandType = "boot", B = 1000, conf = 0.95, detrend = FALSE, ... )
Arguments
| mapping | Set of aesthetic mappings created by |
|---|---|
| data | The data to be displayed in this layer. There are three options: If A A |
| geom | The geometric object to use display the data |
| position | Position adjustment, either as a string, or the result of a call to a position adjustment function. |
| na.rm | If |
| show.legend | logical. Should this layer be included in the legends?
|
| inherit.aes | If |
| distribution | Character. Theoretical probability distribution function
to use. Do not provide the full distribution function name (e.g.,
|
| dparams | List of additional parameters passed on to the previously
chosen |
| bandType | Character. Only |
| B | Integer. If |
| conf | Numerical. Confidence level of the bands. |
| detrend | Logical. Should the plot objects be detrended? If |
| ... | Other arguments passed on to |
Examples
# generate random Normal data set.seed(0) smp <- data.frame(norm = rnorm(100), exp = rexp(100)) # Normal P-P plot of Normal data gg <- ggplot(data = smp, mapping = aes(sample = norm)) + stat_pp_band() + stat_pp_line() + stat_pp_point() + labs(x = "Probability Points", y = "Cumulative Probability") gg# Shifted Normal P-P plot of Normal data dp <- list(mean = 1.5) gg <- ggplot(data = smp, mapping = aes(sample = norm)) + stat_pp_band(dparams = dp) + stat_pp_line() + stat_pp_point(dparams = dp) + labs(x = "Probability Points", y = "Cumulative Probability") gg# Exponential P-P plot of Exponential data di <- "exp" gg <- ggplot(data = smp, mapping = aes(sample = exp)) + stat_pp_band(distribution = di) + stat_pp_line() + stat_pp_point(distribution = di) + labs(x = "Probability Points", y = "Cumulative Probability") gg# Normal P-P plot of mean ozone levels (airquality dataset) dp <- list(mean = 38, sd = 27) gg <- ggplot(data = airquality, mapping = aes(sample = Ozone)) + stat_pp_band(dparams = dp) + stat_pp_line() + stat_pp_point(dparams = dp) + labs(x = "Probability Points", y = "Cumulative Probability") gg